24 February, 2021
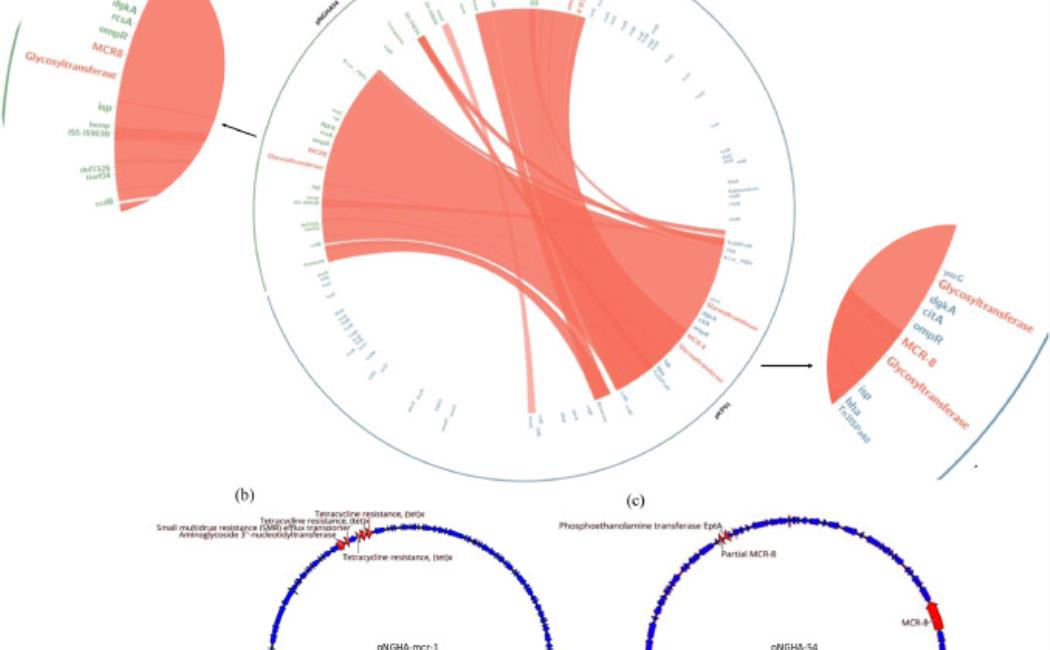
Polymyxin E (colistin) is among the last-resort antibiotics used to treat carbapenem-resistant Enterobacteriaceae-related infections [1]. Colistin had been discontinued for human use since it was found to be associated with nephrotoxicity and neurotoxicity, however, since the 1990s, clinicians were required to reconsider the clinical value of a modified version as a last resort antibiotic. Currently, single plasmid-mediated mobile colistin resistance (mcr) gene is recognized as a global threat, but multiple mcr gene combinations in the same pathogen are rarely identified [2]. Membrane binding domains in MCR proteins are necessary for membrane charge modifications involved in the colistin resistance mechanism [3]. Wang et al. discussed the first mcr-8 containing isolate of Klebsiella pneumoniae (KP) with a high minimum inhibitory concentration (MIC) to colistin in a Chinese hospital in 2016, which was prior to the discovery of the mcr-8 gene in 2017 from a pig farm in China related to colistin usage in animals [3]. Typically, mcr-variants are investigated through molecular identification tools or next-generation sequencing (NGS), which is not routinely implemented in clinical molecular biology laboratories. Such studies identified a close evolutionary relationship between mcr and phosphoethanolamine transferase (eptA) of Neisseria that is known to induce colistin resistance [3]. Multiple variants of mcr genes including co-occurrences in the same bacterial strain in the clinic has been identified through epidemiological surveillance of disease-causing pathogens. Continuous monitoring and identification of clinical cases that harbor unique mcr-variants is essential for understanding and monitoring colistin resistant bacteria. In this study, we identified a unique combination of colistin-resistance genes in a multidrug-resistant (MDR) KP clinical isolate and explored the role of the variant mcr genes in colistin resistance.